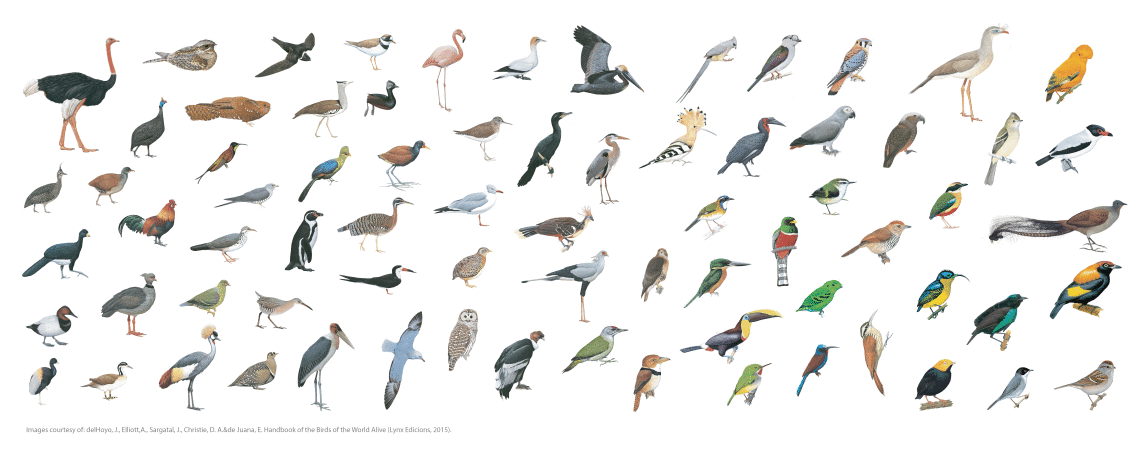
Nature Paper Resolves Bird Tree of Life
The first Anchored Phylogenomics project in birds resolves one of the greatest challenges in dinosaur systematics: resolving the deep branches of the avian phylogeny. The combination of high taxon sampling and high levels of phylogenetic information in anchored loci facilitated identification of several novel clades and successfully placed enigmatic species.
Contact us to request a free PDF if you cannot access the paper from the Nature website below.
2015 Prum et al.
NATURE; doi:10.1038/nature15697 Show Details
5278 Downloads

Largest Amphibian Tree of Life data set presented at SSAR conference
Paul Hime and David Weisrock presented preliminary results from a 300+ taxon data set for hundreds of anchored loci at the Society for the Study of Amphibians and Reptiles 2015 Conference. Early results suggest an unexpected sister relationship between salamanders and caecilians.

Roots of Madagascar snake radiation elucidated
Anchored Phylogenomic data provide support for a monophyletic radiation of Pseudoxyrhophiine snakes in Madagascar. Feeding specialization associated with tooth venom delivery is thought to have played a major role in the early diversification of this radiation.
2015 Ruane et al.
In Press Show Details
2880 Downloads
Hierarchical Anchored Phylogenomics highlighted in symposium
Emily Lemmon presented a new expansion of Anchored Phylogenomics, which facilitates simultaneous data collection for phylogenetics, phylogeography, and population genomics, at the Frontiers in Biology Symposium at the Society for the Study of Amphibians and Reptiles 2015 Conference.

Breaking ground in fish phylogenomics
Improved modeling and Anchored Phylogenomic data facilitate resolution of deep relationships in the fish clade Ovalentaria. This study collected data using the original Vertebrate v1 capture probe design. We have since developed a much improved fish-specific design that have been used in several projects currently underway.

Scincid lizard phylogeny resolved at shallow and deep levels
Anchored Phylogenomics provided resolution at both phylogeographic and phylogenetic levels in skinks. In a comparison to transcriptome data, Anchored Phylogenomic data sets were found to perform as well or better and to be more cost effective, thus allowing greater taxon sampling. This study collected data using the original Vertebrate v1 capture probe design. We have since developed a much improved amniote-specific design.

Science Features Anchored Phylogenomics
The Lemmons and several collaborators were interviewed in a recent article discussing the impact of this method and other hybrid enrichment approaches on the field of phylogenetics.
[button url=”http://www.sciencemag.org” target=”blank”]Access The Article[/button]
The Robot Has Arrived
FSU invests in Anchored Phylogenomics by purchasing a state-of-the-art liquid-handling robot capable of processing 25,000 samples per year. Funding for the Beckman-Coulter Biomek FXp was provided by Florida State University.
Annual Reviews Article Now Online
Our paper reviewing the effect of high-throughput genomics on data collection and analysis in phylogenetics is now published online in Annual Review of Ecology, Evolution, and Systematics.
[button url=”http://anchoredphylogeny.com/publications/” size=”4″]Publications[/button]Anchored Hybrid Enrichment for Massively High-Throughput Phylogenomics
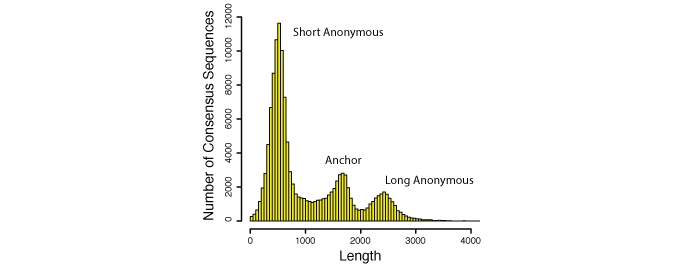
The field of phylogenetics is on the cusp of a major revolution, enabled by new methods of data collection that leverage both genomic resources and recent advances in DNA sequencing. Previous phylogenetic work has required labor-intensive marker development coupled with single-locus polymerase chain reaction and DNA sequencing on clade-by-clade and locus-by-locus basis. Here, we present a new, cost-efficient, and rapid approach to obtaining data from hundreds of loci for potentially hundreds of individuals for deep and shallow phylogenetic studies. Specifically, we designed probes for target enrichment of >500 loci in highly conserved anchor regions of vertebrate genomes (flanked by less conserved regions) from five model species and tested enrichment efficiency in nonmodel species up to 508 million years divergent from the nearest model.
We found that hybrid enrichment using conserved probes (anchored enrichment) can recover a large number of unlinked loci that are useful at a diversity of phylogenetic timescales. This new approach has the potential not only to expedite resolution of deep-scale portions of the Tree of Life but also to greatly accelerate resolution of the large number of shallow clades that remain unresolved. The combination of low cost (∼1% of the cost of traditional Sanger sequencing and ∼3.5% of the cost of high-throughput amplicon sequencing for projects on the scale of 500 loci × 100 individuals) and rapid data collection (∼2 weeks of laboratory time) are expected to make this approach tractable even for researchers working on systems with limited or nonexistent genomic resources.
2012 Lemmon, A. R., S. Emme and E. C. Lemmon
SYSTEMATIC BIOLOGY. 61: 721-744. Show Details
4001 Downloads